

植物生态学报 ›› 2006, Vol. 30 ›› Issue (3): 450-456.DOI: 10.17521/cjpe.2006.0060
所属专题: 碳水能量通量
接受日期:2005-09-08
发布日期:2006-05-30
通讯作者:
朱波
作者简介:*E-mail: bzhu@imde.ac.cn基金资助:
HAN Guang-Xuan1,2( ), ZHU Bo2,*, JIANG Chang-Sheng3
), ZHU Bo2,*, JIANG Chang-Sheng3
Accepted:2005-09-08
Published:2006-05-30
Contact:
ZHU Bo
摘要:
基于川中丘陵区2003年4~9月水稻田土壤呼吸、土壤温度和水稻(Oryza sativa)生物量的测定,研究了水稻田土壤呼吸日变化和季节变化特征以及影响稻田土壤呼吸的主要因素。结果表明,水稻田土壤CO2排放通量的日变化为单峰型,其最小值和最大值分别出现在当地时间7∶00和15∶00;在水稻生长期内,稻田土壤CO2排放通量在18.00~269.69 mg·m-2·h-1之间波动,平均排放通量为121.76 mg·m-2·h-1。在日的时间尺度上,水稻田土壤CO2排放通量与5 cm土壤温度存在显著的指数函数关系;而从整个生长期时间尺度上看,水稻田土壤CO2的排放通量主要受到5 cm土壤温度和水稻地下生物量的影响。在水稻生长初期,水稻地下生物量与稻田土壤CO2排放通量之间存在着显著的相关关系;水稻拔节中后期到成熟期,土壤温度则是制约稻田土壤CO2排放的关键因素。CO2排放通量与稻田地表水层深度的相关关系不显著。
韩广轩, 朱波, 江长胜. 川中丘陵区水稻田土壤呼吸及其影响因素. 植物生态学报, 2006, 30(3): 450-456. DOI: 10.17521/cjpe.2006.0060
HAN Guang-Xuan, ZHU Bo, JIANG Chang-Sheng. SOIL RESPIRATION AND ITS CONTROLLING FACTORS IN RICE FIELDS IN THE HILL REGION OF THE CENTRAL SICHUAN BASIN. Chinese Journal of Plant Ecology, 2006, 30(3): 450-456. DOI: 10.17521/cjpe.2006.0060
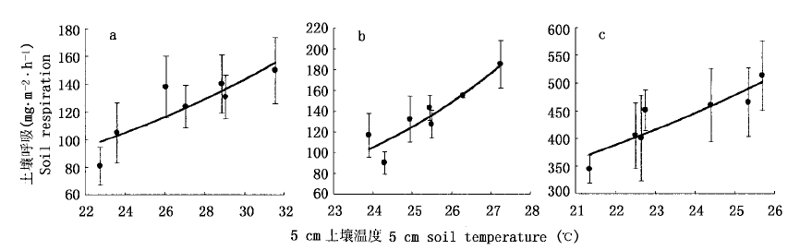
图2 稻田土壤呼吸与5 cm土壤温度的相关关系 a:5月28日 May 28 b:7月22日 July 22 c:9月5日 September 5
Fig.2 Relationship between soil respiration from rice field and 5 cm soil temperature
| 日期 Date | 5 cm土壤温度 5 cm soil temperature | 地表温度 Surface ground temperature | 箱内温度 Air temperature in chamber | 气温 Air temperature |
|---|---|---|---|---|
| 5月28日 May 28 | 0.887** | 0.881** | 0.953** | 0.930** |
| 7月22日 July 22 | 0.913** | 0.672 | 0.752 | 0.743 |
| 9月5日 September 5 | 0.914** | 0.395 | 0.678 | 0.522 |
表1 土壤呼吸与各温度之间的相关关系
Table 1 Correlation relationship between soil respiration and different temperatures
| 日期 Date | 5 cm土壤温度 5 cm soil temperature | 地表温度 Surface ground temperature | 箱内温度 Air temperature in chamber | 气温 Air temperature |
|---|---|---|---|---|
| 5月28日 May 28 | 0.887** | 0.881** | 0.953** | 0.930** |
| 7月22日 July 22 | 0.913** | 0.672 | 0.752 | 0.743 |
| 9月5日 September 5 | 0.914** | 0.395 | 0.678 | 0.522 |
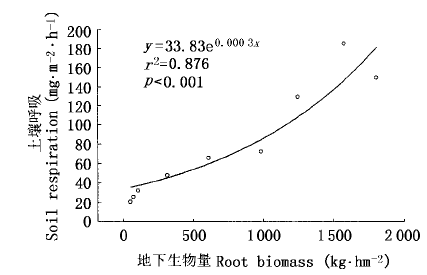
图5 稻田土壤呼吸与水稻地下生物量的关系(自移栽到移栽后35 d)
Fig.5 Relationship between root biomass and soil respiration from rice field (from transplanting to 35 days after transplanting)
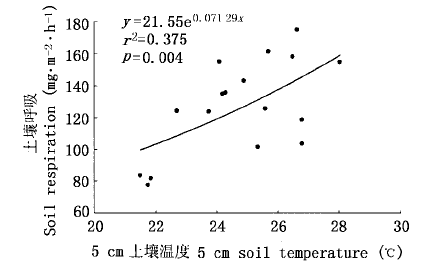
图7 稻田土壤呼吸与5 cm土壤温度的关系(自移栽后42 d到收获)
Fig.7 Relationship between 5 cm soil temperature and soil respiration from rice field (from 42 days after transplanting to reaping)
| [1] | Bouma TJ, Bryla DR (2000). On the assessment of root and soil respiration for soils of different textures: interactions with soil moisture contents and soil CO2 concentrations . Plant and Soil, 227,215-221. |
| [2] | Bouma TJ, Nielsen KL, Eissenstat DM, Lynch JP (1997). Estimating respiration of roots in soil: interactions with soil CO2, soil temperature, and soil water content . Plant and Soil, 195,221-232. |
| [3] |
Burton AJ, Pregitzer KS (2003). Field measurements of root respiration indicate little to no seasonal temperature acclimation for sugar maple and red pine. Tree physiology, 23,273-280.
URL PMID |
| [4] | Davidson EA, Belk E, Boone RD (1998). Soil water content and temperature as independent or confounded factors controlling soil respiration in a temperate mixed hardwood forest. Global Change Biology, 4,217-227. |
| [5] |
Davidson EA, Trumbore SE, Amundson R (2000). Soil warming and organic carbon content. Nature, 408,789-790.
DOI URL PMID |
| [6] | Elberling B (2003). Seasonal trends of soil CO2 dynamics in a soil subject to freezing . Journal of Hydrology, 276,159-175. |
| [7] | Fang C, Moncrieff JB (1999). A model for soil CO2 production and transport. I. Model development . Agricultural and Forest Meteorology, 95,225-236. |
| [8] | Fang C, Moncrieff JB (2001). The dependence of soil CO2 efflux on temperature . Soil Biology and Biochemistry, 33,155-165. |
| [9] | Frank AB, Liebig MA, Hanson JD (2002). Soil carbon dioxide fluxes in northern semiarid grasslands. Soil Biology and Biochemistry, 34,1235-1241. |
| [10] | Gough M, Seiler JR (2004). The influence of environmental, soil carbon, root, and stand characteristics on soil CO2 efflux in loblolly pine ( Pinus taeda L.) plantations located on the South Carolina Coastal Plain Christopher . Forest Ecology and Management, 191,353-363. |
| [11] |
Grace J, Rayment M (2000). Respiration in the balance. Nature, 404,819-820.
DOI URL PMID |
| [12] | Hanson PJ, Edwards NT, Garten CT, Andrews JA (2000). Separating root and soil microbial contributions to soil respiration: a review of methods and observations. Biogeochemistry, 48,115-146. |
| [13] | IPCC (2001). Climate Change 2001. Third Assessment Report of the IPCC. Cambridge University Press, Cambridge,183-237. |
| [14] | Jenkinson DS, Adams DE, Wild A (1991). Model estimates of CO2 emission from soil in response to global warming . Nature, 351,304-306. |
| [15] | Liang W (梁巍), Yue J (岳进), Shi Y (史奕), Huang GH (黄国宏), Liang ZB (梁战备) (2003). Seasonal variation of soil microbial biomass, respiration rate and CH4 emission in black earth rice fields . Chinese Journal of Applied Ecology (应用生态学报), 14,2278-2280. (in Chinese with English abstract) |
| [16] | Maier CA, Kress LW (2000). Soil CO2 evolution and root respiration in 11-year-old loblolly pine ( Pinus taeda) plantations as affected by moisture and nutrient availability . Canadian Journal of Forest Research, 30,347-359. |
| [17] | Mariko S, Nishimura N, Mo W (2000). Winter CO2 flux from soil and snow surfaces in a cool-temperate deciduous forest . Ecological Research, 15,363-372. |
| [18] | Pangle RE, Seiler J (2002). Influence of seedling roots, environmental factors and soil characteristics on soil CO2 efflux rates in a 2-year-old loblolly pine ( Pinus taeda L.) plantation in the Virginia Piedmont . Environmental Pollution, 116,s85-s96. |
| [19] | Raich JW, Schlesinger WH (1992). The global carbon dioxide flux in soil respiration to vegetation. Tellus, 44(B),81-99. |
| [20] | Richard DB, Eric D, Kathleen S (2004). Chronic nitrogen additions reduce total soil respiration and microbial respiration in temperate forest soils at the Harvard Forest. Forest Ecology and Management, 196,43-56. |
| [21] | Rustad LE, Huntington TG, Boone RD (2000). Controls on soil respiration: implications for climate change. Biogeochemistry, 49,1-6. |
| [22] | Schimel DS (1995). Terrestrial ecosystem and carbon-cycle. Global Change Biology, 1,77~99 . |
| [23] | Sun WJ (孙文娟), Huang Y (黄耀), Chen ST (陈书涛), Yang ZF (杨兆芳), Zheng XH (郑循华) (2004). CO2 emission from soil-crop system as influenced by crop growth and tissue N content . Environmental Science (环境科学), 25(3),1-6. (in Chinese with English abstract) |
| [24] | Tang JW, Dennis DB, Qi Y, Xu LK (2003). Assessing soil CO2 efflux using continuous measurements of CO2 profiles in soils with small solid-state sensors . Agricultural and Forest Meteorology, 118,207-220. |
| [25] | Treonis AM, Wall DH, Virginia RA (2002). Field and microcosm studies of decomposition and soil biota in a cold desert soil. Ecosystems, 5,159-170. |
| [26] | Wang ZF (王子芳), Gao M (高明), Qin JC (秦建成), Ci E (慈恩) (2003). Effect of long-term “paddy-upland” rotation on soil fertility of paddy fields. Journal of Southwest Agricultural University(Natural Science Edition) (西南农业大学学报(自然科学版)), 25,514-521. (in Chinese with English abstract) |
| [27] | Wiseman PH, Seiler JR (2004). Soil CO2 efflux across four age classes of plantation loblolly pine ( Pinus taeda L.) on the Virginia Piedmont . Forest Ecology and Management, 192,297-311. |
| [28] | Yu GR (于贵瑞) (2003). Global Change, Carbon Cycle and Storage in Terrestrial Ecosystem (全球变化与陆地生态系统碳循环和碳蓄积). China Meteorological Press, Beijing,1-4. (in Chinese) |
| [29] |
Yuste JC, Janssens IA, Carrara A, Meiresonne L, Ceulemans R (2003). Interactive effects of temperature and precipitation on soil respiration in a temperate maritime pine forest. Tree Physiology, 23,1263-1270.
DOI URL PMID |
| [30] | Zhou ZT (周志田), Cheng SK (成升魁), Liu YF (刘允芬), Li JY (李家永) (2002). CO2 emission of soil under different land-use types in subtropical red soil hilly areas in China: preliminary exploration . Resources Science (资源科学), 24(2),83-87. (in Chinese with English abstract) |
| [31] | Zou JW (邹建文), Huang Y (黄耀), Zong LG (宗良纲), Zheng XH (郑循华), Wang YS (王跃思) (2003). A field study on CO2, CH4 and N2O emissions from rice paddy and impact factors . Acta Scientiae Circumstantiae (环境科学学报), 23,758-764. (in Chinese with English abstract) |
| [1] | 沈健, 何宗明, 董强, 郜士垒, 林宇. 轻度火烧对滨海沙地人工林土壤呼吸速率和非生物因子的影响[J]. 植物生态学报, 2023, 47(7): 1032-1042. |
| [2] | 杨元合, 张典业, 魏斌, 刘洋, 冯雪徽, 毛超, 徐玮婕, 贺美, 王璐, 郑志虎, 王媛媛, 陈蕾伊, 彭云峰. 草地群落多样性和生态系统碳氮循环对氮输入的非线性响应及其机制[J]. 植物生态学报, 2023, 47(1): 1-24. |
| [3] | 郑甲佳, 黄松宇, 贾昕, 田赟, 牟钰, 刘鹏, 查天山. 中国森林生态系统土壤呼吸温度敏感性空间变异特征及影响因素[J]. 植物生态学报, 2020, 44(6): 687-698. |
| [4] | 杨泽, 嘎玛达尔基, 谭星儒, 游翠海, 王彦兵, 杨俊杰, 韩兴国, 陈世苹. 氮添加量和施氮频率对温带半干旱草原土壤呼吸及组分的影响[J]. 植物生态学报, 2020, 44(10): 1059-1072. |
| [5] | 胡姝娅,刁华杰,王惠玲,薄元超,申颜,孙伟,董宽虎,黄建辉,王常慧. 北方农牧交错带温性盐碱化草地土壤呼吸对不同形态氮添加和刈割的响应[J]. 植物生态学报, 2020, 44(1): 70-79. |
| [6] | 温超,单玉梅,晔薷罕,张璞进,木兰,常虹,任婷婷,陈世苹,白永飞,黄建辉,孙海莲. 氮和水分添加对内蒙古荒漠草原放牧生态系统土壤呼吸的影响[J]. 植物生态学报, 2020, 44(1): 80-92. |
| [7] | 王明明,刘新平,何玉惠,张铜会,魏静,车力木格,孙姗姗. 科尔沁沙地封育恢复过程中植物群落特征变化及影响因素[J]. 植物生态学报, 2019, 43(8): 672-684. |
| [8] | 刘程竹, 贾娟, 戴国华, 马田, 冯晓娟. 中性糖在土壤中的来源与分布特征[J]. 植物生态学报, 2019, 43(4): 284-295. |
| [9] | 皮春燕, 刘鑫, 王喆, 包维楷. 苔藓-蓝藻共生体关系与固氮能力研究进展[J]. 植物生态学报, 2018, 42(4): 407-418. |
| [10] | 王祥, 朱亚琼, 郑伟, 关正翾, 盛建东. 昭苏山地草甸4种典型土地利用方式下的土壤呼吸特征[J]. 植物生态学报, 2018, 42(3): 382-396. |
| [11] | 杨开军, 杨万勤, 谭羽, 贺若阳, 庄丽燕, 李志杰, 谭波, 徐振锋. 川西亚高山云杉林冬季土壤呼吸对雪被去除的短期响应[J]. 植物生态学报, 2017, 41(9): 964-971. |
| [12] | 李茜, 王芳, 曹扬, 彭守璋, 陈云明. 陕西省森林土壤固碳特征及其影响因素[J]. 植物生态学报, 2017, 41(9): 953-963. |
| [13] | 郭建荣, 万贤崇. 杨树根压昼夜周期性及其影响因子[J]. 植物生态学报, 2017, 41(3): 369-377. |
| [14] | 杨青霄, 田大栓, 曾辉, 牛书丽. 降水格局改变背景下土壤呼吸变化的主要影响因素及其调控过程[J]. 植物生态学报, 2017, 41(12): 1239-1250. |
| [15] | 葛晓改, 周本智, 肖文发, 王小明, 曹永慧, 叶明. 生物质炭添加对毛竹林土壤呼吸动态和温度敏感性的影响[J]. 植物生态学报, 2017, 41(11): 1177-1189. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19